Watch Now This tutorial has a related video course created by the Real Python team. Watch it together with the written tutorial to deepen your understanding: NumPy Techniques and Practical Examples
The NumPy library is a Python library used for scientific computing. It provides you with a multidimensional array object for storing and analyzing data in a wide variety of ways. In this tutorial, you’ll see examples of some features NumPy provides that aren’t always highlighted in other tutorials. You’ll also get the chance to practice your new skills with various exercises.
In this tutorial, you’ll learn how to:
- Create multidimensional arrays from data stored in files
- Identify and remove duplicate data from a NumPy array
- Use structured NumPy arrays to reconcile the differences between datasets
- Analyze and chart specific parts of hierarchical data
- Create vectorized versions of your own functions
If you’re new to NumPy, it’s a good idea to familiarize yourself with the basics of data science in Python before you start. Also, you’ll be using Matplotlib in this tutorial to create charts. While it’s not essential, getting acquainted with Matplotlib beforehand might be beneficial.
Get Your Code: Click here to download the free sample code that you’ll use to work through NumPy practical examples.
Take the Quiz: Test your knowledge with our interactive “NumPy Practical Examples: Useful Techniques” quiz. You’ll receive a score upon completion to help you track your learning progress:
Interactive Quiz
NumPy Practical Examples: Useful TechniquesThis quiz will test your understanding of working with NumPy arrays. You won't find all the answers in the tutorial, so you'll need to do some extra investigating. By finding all the answers, you're sure to learn some interesting things along the way.
Setting Up Your Working Environment
Before you can get started with this tutorial, you’ll need to do some initial setup. In addition to NumPy, you’ll need to install the Matplotlib library, which you’ll use to chart your data. You’ll also be using Python’s pathlib library to access your computer’s file system, but there’s no need to install pathlib because it’s part of Python’s standard library.
You might consider using a virtual environment to make sure your tutorial’s setup doesn’t interfere with anything in your existing Python environment.
Using a Jupyter Notebook within JupyterLab to run your code instead of a Python REPL is another useful option. It allows you to experiment and document your findings, as well as quickly view and edit files. The downloadable version of the code and exercise solutions are presented in Jupyter Notebook format.
The commands for setting things up on the common platforms are shown below:
You’ll notice that your prompt is preceded by (venv). This means that anything you do from this point forward will stay in this environment and remain separate from other Python work you have elsewhere.
Now that you have everything set up, it’s time to begin the main part of your learning journey.
NumPy Example 1: Creating Multidimensional Arrays From Files
When you create a NumPy array, you create a highly-optimized data structure. One of the reasons for this is that a NumPy array stores all of its elements in a contiguous area of memory. This memory management technique means that the data is stored in the same memory region, making access times fast. This is, of course, highly desirable, but an issue occurs when you need to expand your array.
Suppose you need to import multiple files into a multidimensional array. You could read them into separate arrays and then combine them using np.concatenate(). However, this would create a copy of your original array before expanding the copy with the additional data. The copying is necessary to ensure the updated array will still exist contiguously in memory since the original array may have had non-related content adjacent to it.
Constantly copying arrays each time you add new data from a file can make processing slow and is wasteful of your system’s memory. The problem becomes worse the more data you add to your array. Although this copying process is built into NumPy, you can minimize its effects with these two steps:
-
When setting up your initial array, determine how large it needs to be before populating it. You may even consider over-estimating its size to support any future data additions. Once you know these sizes, you can create your array upfront.
-
The second step is to populate it with the source data. This data will be slotted into your existing array without any need for it to be expanded.
Next, you’ll explore how to populate a three-dimensional NumPy array.
Populating Arrays With File Data
In this first example, you’ll use the data from three files to populate a three-dimensional array. The content of each file is shown below, and you’ll also find these files in the downloadable materials:
The first file has two rows and three columns with the following content:
file1.csv
1.1, 1.2, 1.3
1.4, 1.5, 1.6
This second file, which also has the same dimensions, contains this:
file2.csv
2.1, 2.2, 2.3
2.4, 2.5, 2.6
The third file, again with the same dimensions, stores these numbers:
file3.csv
3.1, 3.2, 3.3
3.4, 3.5, 3.6
Before you continue, add these three files to your program folder. The downloadable materials also contain two files called file10.csv and file11.csv, and you’ll be working with these later on.
The diagram below shows the resulting NumPy array you’ll create from the three files:
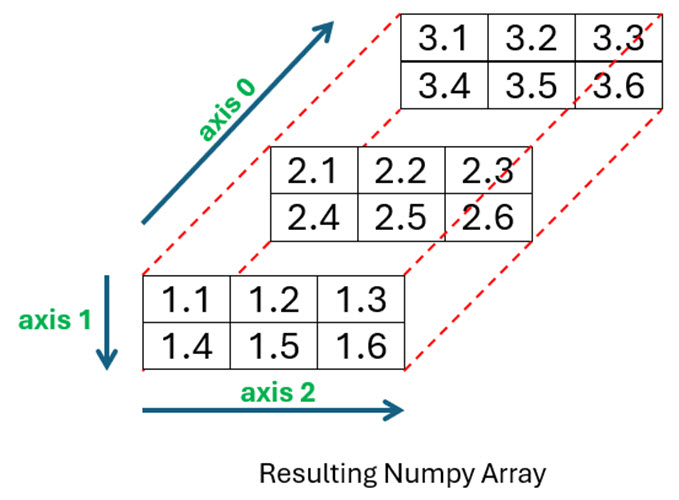
As you can see, file1.csv forms the front of your array, file2.csv the middle section, and file3.csv is at the back.
The code used to create this array is shown below. Before you run this code, make sure you either create the three files shown in the diagram or use the versions provided in the downloadable materials. Either way, place them in the same directory you’re running your code in, then run it:
1>>> from pathlib import Path
2>>> import numpy as np
3
4>>> array = np.zeros((3, 2, 3))
5>>> print(id(array))
62250027286128
7
8>>> for file_count, csv_file in enumerate(Path.cwd().glob("file?.csv")):
9... array[file_count] = np.loadtxt(csv_file.name, delimiter=",")
10...
11>>> print(id(array))
122250027286128
13
14>>> print(array.shape)
15(3, 2, 3)
16
17>>> array
18array([[[1.1, 1.2, 1.3],
19 [1.4, 1.5, 1.6]],
20
21 [[2.1, 2.2, 2.3],
22 [2.4, 2.5, 2.6]],
23
24 [[3.1, 3.2, 3.3],
25 [3.4, 3.5, 3.6]]])
To begin, you can look through each of the files and use the information to determine the final shape of the expected array. In this example, all three files have the same number of elements arranged in two rows and three columns. The resulting array will then have a shape property of (3, 2, 3). Take another look at the diagram and you’ll be able to see this.
Lines 1 and 2 show the NumPy library being imported with its standard alias of np and also the Path class being imported from the pathlib library. This library allows Python to access your computer’s file system using an object-oriented approach. Objects of the Path class allow you to specify a file path and also contain methods so you can make system calls to your operating system. This feature is used later in the code.
You learned earlier how it’s a good idea to create an array upfront before you start populating it with data to reduce the memory footprint. In line 4, you create an array containing zeros with a shape of (3, 2, 3) as you determined earlier.
Next, you populate your array with the data from the files. You create a for loop using Python’s built-in enumerate function shown in lines 8 and 9. Its parameter allows you to loop over a set of files and return a reference to each one. It also maintains a count of how many files have been encountered. Each file reference is stored in the csv_file variable, while the incrementing counter is stored in the file_count variable.
To access each of the three .csv files in turn, you use Path. By calling Path.cwd(), you’re telling Python to look in your current working directory for the files. In other words, the directory from where you’re running the program. This returns a Path object representing the current directory, from which you call its .glob() method to specify the names of the files you wish to access.
In this case, because you want to access files named file1.csv, file2.csv, and file3.csv, you pass their names in as the string file?.csv. This tells .glob() to select only files whose names must match these exact characters, but whose fifth character can be any character, as specified by the wildcard character (?).
Unfortunately, .glob() may not return the files in the order you expect. In this example, everything does work as expected because each file’s name contains a single digit as its fifth character. If there had been a file named file11.csv, then it would have been read in the wrong order. You’ll learn more about why this is and how to solve it later.
Note: When using Windows, .glob() will yield the files sorted lexicographically. That’s not necessarily the case on other operating systems. If you rely on the files being sorted, you should explicitly call sorted() as well:
>>> for csv_file in sorted(Path.cwd().glob("file?.csv")):
... print(csv_file.name)
...
file1.csv
file2.csv
file3.csv
As you’ll see, this kind of sorting is not always enough when working with numbered files.
Each time the loop loops, you call the np.loadtxt() function and pass it a filename, which is specified using its name property. You also tell it to use a comma (,) as a field delimiter to allow it to separate out each individual number in the file. The content of each file is then assigned to the array you created earlier.
To make sure the content of each file is inserted into the correct position along the 0 axis, you use array[file_count]. The first time the loop executes, the contents of file1.csv will be assigned to array[0], or position 0 along axis-0. The next iteration in the loop will assign file2.csv to array[1] along this axis, before file3.csv gets assigned to array[2]. Look once more at the diagram and you’ll see exactly what’s happened.
In lines 5 and 11, you printed the result of id(array). The id() function returns the identity of an object. Each object has a unique identity value because each object occupies a unique place in computer memory. When you run the code on your computer, the numbers will also be identical to each other, but will probably be different from those shown.
In lines 6 and 12, the identity values displayed prove that the array object that started off as containing only zeros, is the same array object that later contained the contents of each file. This shows that only one object was used throughout and that memory was used efficiently.
When you create arrays in this way, it’s a good idea to make sure that each of the input files has the same number of rows and columns of elements. Next, you’ll look at how to deal with situations where your data files are not quite so uniform.
Dealing With Different Data Sizes
To begin with, you’ll add in some undersized data. You’ll find it in a file named short_file.csv, which only has a single row:
short_file.csv
4.1, 4.2, 4.3
You want to add it to the back of your array as shown below:
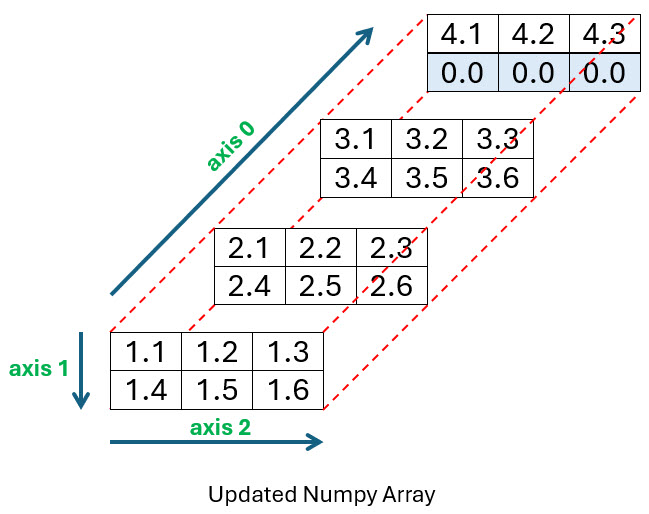
Before you run the code used to create this second array, make sure you either download or add the file named short_file.csv into the same directory you run your code:
1>>> array = np.zeros((4, 2, 3))
2
3>>> for file_count, csv_file in enumerate(Path.cwd().glob("file?.csv")):
4... array[file_count] = np.loadtxt(csv_file.name, delimiter=",")
5...
6>>> array[3, 0] = np.loadtxt("short_file.csv", delimiter=",")
7
8>>> array
9array([[[1.1, 1.2, 1.3],
10 [1.4, 1.5, 1.6]],
11
12 [[2.1, 2.2, 2.3],
13 [2.4, 2.5, 2.6]],
14
15 [[3.1, 3.2, 3.3],
16 [3.4, 3.5, 3.6]],
17
18 [[4.1, 4.2, 4.3],
19 [0. , 0. , 0. ]]])
This time, you’re reading in four separate files, so the array you initially create will have a shape of (4, 2, 3). You’ll need an extra dimension along axis-0 to accommodate the fourth file, so you create this in line 1.
The for loop is used to read the first three files as before. To read in the short file, you need to tell Python exactly where you want to place it in your array. Previously, you did this by indicating a position, such as array[2], to insert the data into position 2 along axis-0. This approach worked because the data you inserted filled the existing array already at that position. However, this time things are different.
To tell Python you want to insert the short file starting at the top of index position 3 in axis-2, you use array[3, 0]. The 3 represents the axis-2 position as before, but you must also supply a 0 to indicate that the data should be inserted starting at row 0. You can take a look at lines 18 and 19 of the program output and then at the diagram to see where the data was placed.
As before, the array object created at the start of the code is the only one used throughout. Although your code added data to it several times, no inefficient copying was needed because you created the array upfront.
Suppose that instead of being too short, the fourth file was too long. You may be wondering how to deal with such files, which can be problematic. This time, you’ll use a file named long_file.csv, which has an extra row:
long_file.csv
4.1, 4.2, 4.3
4.4, 4.5, 4.6
4.7, 4.8, 4.9
Now you want to incorporate it into your array at the position shown below. As you can see from the diagram, the rest of the array will need to be extended by an extra row to accommodate it:
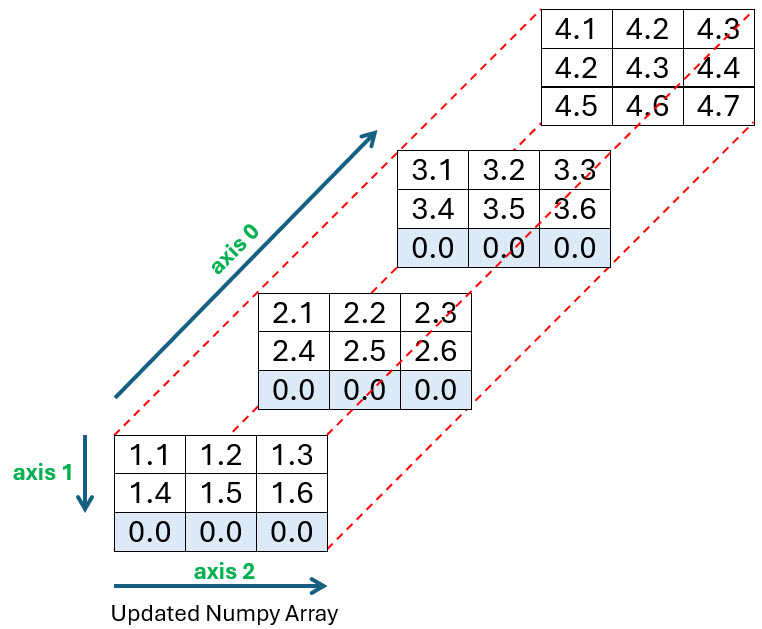
The code used to create this third array is shown below. Before you run it, make sure to either download or create the file named long_file.csv in the same directory you’re running your code:
1>>> array = np.zeros((4, 2, 3))
2
3>>> print(id(array))
42250027278352
5
6>>> for file_count, csv_file in enumerate(Path.cwd().glob("file?.csv")):
7... array[file_count] = np.loadtxt(csv_file.name, delimiter=",")
8...
9>>> array = np.insert(arr=array, obj=2, values=0, axis=1)
10
11>>> array[3] = np.loadtxt("long_file.csv", delimiter=",")
12
13>>> print(id(array))
142250027286224
15
16>>> array
17array([[[1.1, 1.2, 1.3],
18 [1.4, 1.5, 1.6],
19 [0. , 0. , 0. ]],
20
21 [[2.1, 2.2, 2.3],
22 [2.4, 2.5, 2.6],
23 [0. , 0. , 0. ]],
24
25 [[3.1, 3.2, 3.3],
26 [3.4, 3.5, 3.6],
27 [0. , 0. , 0. ]],
28
29 [[4.1, 4.2, 4.3],
30 [4.4, 4.5, 4.6],
31 [4.7, 4.8, 4.9]]])
This time, because your original array is too short to accommodate the contents of long_file.csv, you need to lengthen it with an extra row along axis-1. You can then add in the content of long_file.csv. You do this in line 9 by using the np.insert() function, which is a function that allows you to insert values along an axis.
You pass four parameters to np.insert(). The arr parameter specifies the array you want to insert values into, while setting obj to 2, axis to 1, and the values parameter to 0, allows you to insert 0 values into index position 2 along axis 1. In other words, along the bottom row of the array as shown in the diagram. Finally, to add the content of long_file.csv into your array, you once more use loadtxt() as shown in line 11.
Take a moment to look at the diagram and the resulting array produced by your code and you’ll see that everything works as expected.
Notice that lines 4 and 14 indicate that the array objects before and after the new data is inserted are different. This occurs because the insert() function returns a copy of the original array. To avoid this waste of memory, it really is a good idea to make sure you size your initial array correctly before you start populating it.
Making Sure File Order Is Correct
When you run the Path.cwd().glob("file?.csv") code, you return a generator iterator that can be used to view a collection of WindowsPath or PosixPath objects. Each of these represents an operating system file path and filename that matches the file?.csv pattern. However, the order in which these files are returned by the generator isn’t what you might expect.
To see an example of this, add the two files named file10.csv and file11.csv from the downloadable materials into your existing folder:
file10.csv
10.1,10.2,10.3
10.4,10.5,10.6
You probably already guessed what was in file10.csv. If so, you won’t be surprised when you see what’s in file11.csv:
file11.csv
11.1,11.2,11.3
11.4,11.5,11.6
Now run the following code:
>>> for csv_file in Path.cwd().glob("file*.csv"):
... print(csv_file.name)
...
file1.csv
file10.csv
file11.csv
file2.csv
file3.csv
To make sure each of these additional files are seen by the generator, you need to adjust the match criteria to file*.csv. The (*) wildcard character represents any number of unknown characters. If you had retained the (?) wildcard character, then only the files containing one character after the string file would have been included. Even with this addition, something still looks wrong.
Note: On Linux and macOS, you may need to add a call to sorted() to see the files ordered as above.
You’ll see that the new files are placed in between file1.csv and file2.csv and not, as you probably expected, at the end. The reason for this is that the filenames are being sorted alphanumerically. This means that the sort reads the filenames from left to right and considers everything equal until it finds a difference. Once it has this, the sort is based on that difference.
For example, when the characters in each filename are being analyzed, everything is considered equal over the first four characters of each file’s name—in this case, file. Python then has to decide which of the fifth characters comes first. It does this by considering the Unicode character code numbers of each of them.
The Unicode character code for the character 1 is 49, while the codes for 2 and 3 are 50 and 51, respectively. As a result, any filename with 1 as its fifth character will then be sorted earlier than those with 2 or 3 in the same position.
In the case of file1.csv, file10.csv, and file11.csv, the fifth character of each filename is the same. Therefore, the sort order will be decided by their sixth character. When considering their Unicode character values, the period (.) has a value of 46, which comes before both the characters 0 and 1, whose values are 48 and 49, respectively. Consequently, the sort order will be file1.csv, followed by file10.csv, and then file11.csv.
You might be tempted to give Python’s built-in sorted() function a try to see if that helps:
>>> for csv_file in sorted(Path.cwd().glob("file*.csv")):
... print(csv_file.name)
...
file1.csv
file10.csv
file11.csv
file2.csv
file3.csv
However, the sorted() function has given you the same undesirable result as before.
To read files in a more natural order, you can use the natsort library. First, install it by running the command python -m pip install natsort. After installation, you can import the natsorted() function and use it instead of the built-in sorted() function to achieve more natural sorting of the files. Below is the code illustrating this:
>>> from natsort import natsorted
>>> for csv_file in natsorted(Path.cwd().glob("file*.csv")):
... print(csv_file.name)
...
file1.csv
file2.csv
file3.csv
file10.csv
file11.csv
At last, you’ve managed to sort out your sorting of filenames issue. You can now take your earlier code a step further and add the file contents into the correct places of your array:
>>> array = np.zeros((5, 2, 3))
>>> for file_count, csv_file in enumerate(
... natsorted(Path.cwd().glob("file*.csv"))
... ):
... array[file_count] = np.loadtxt(csv_file.name, delimiter=",")
...
>>> array
array([[[ 1.1, 1.2, 1.3],
[ 1.4, 1.5, 1.6]],
[[ 2.1, 2.2, 2.3],
[ 2.4, 2.5, 2.6]],
[[ 3.1, 3.2, 3.3],
[ 3.4, 3.5, 3.6]],
[[10.1, 10.2, 10.3],
[10.4, 10.5, 10.6]],
[[11.1, 11.2, 11.3],
[11.4, 11.5, 11.6]]])
This time you pass the various file paths into natsorted(), which will sort them in the way you probably intended. The output shows that the content of both file10.csv and file11.csv are now in their correct places within the arrays. Again, notice how the (*) wildcard operator was used here. Also, the dimensions of this version of array have been increased to (5, 2, 3) to provide space for the new data.
Note: A common trick to ensure that numbered files are handled in order is to pad the numbers with leading zeros. If the files had been named file01.csv, file02.csv, file03.csv, file10.csv, and file11.csv, then sorted() would be able to sort the filenames.
As you can see, it’s perfectly possible to construct NumPy arrays from data in files. However, you do need to make sure different sizes are taken into account.
Before moving on, you’ll complete an exercise to test your understanding. This is the first of several exercises included in this tutorial to help you consolidate your learning.
Testing Your Skills: Populating Arrays With File Data
It’s time for you to test your understanding of creating arrays from file data. See if you can solve your first challenge shown below:
Find the ex1a.csv, ex1b.csv, and ex1c.csv files in the downloadable materials. Now, apply the techniques you’ve learned to create a correctly sized three-dimensional array that will allow each file to be added. The content of each file should be inserted so that it touches the top-left corner of a separate index along the 0-axis. You should insert the contents of the ex1b.csv file as a row, and the contents of ex1c.csv as a column.
One possible solution is:
>>> import numpy as np
>>> from pathlib import Path
>>> array = np.zeros((3, 4, 4), dtype="i8")
>>> array[0] = np.loadtxt("ex1a.csv", delimiter=",")
>>> narrow_array = np.loadtxt("ex1b.csv", delimiter=",")
>>> narrow_array = np.insert(arr=narrow_array, values=0, obj=3, axis=0)
>>> array[1, 0] = narrow_array
>>> short_array = np.loadtxt("ex1c.csv", delimiter=",")
>>> short_array = np.insert(arr=short_array, values=0, obj=3, axis=0)
>>> array[2, :, 0] = short_array
>>> array
array([[[ 5, 10, 15, 20],
[25, 30, 35, 40],
[45, 50, 55, 60],
[65, 70, 75, 80]],
[[ 1, 3, 5, 0],
[ 0, 0, 0, 0],
[ 0, 0, 0, 0],
[ 0, 0, 0, 0]],
[[ 2, 0, 0, 0],
[ 4, 0, 0, 0],
[ 6, 0, 0, 0],
[ 0, 0, 0, 0]]])
First, you look through the files and see that an array with the size (3, 4, 4) is necessary to ensure your data fits. You then create an array of zeros with this size, and specify that it’ll store integers. Next, you assign it to the array variable before populating array[0], or axis-0, with the contents of file ex1a.csv, which fits the shape snugly.
The ex1b.csv file has only three values, meaning it’s too short to be directly inserted into array[1]. To fix this, you first read it into narrow_array and then use np.insert() to add an extra 0 into its index position 3, as defined by its values and obj parameters. Finally, you insert narrow_array into the top row of array at index 1 along its 0-axis using array[1, 0].
The ex1c.csv file also has only three values. This means you must adjust it to fit the column, so you add an extra zero onto the end of it. This time, to add short_array, you use array[2, :, 0]. It’ll be inserted at index position 2 along the 0-axis, but :, 0 means that it’ll be inserted down the first column and not across a row.
Instead of extending the arrays that you read, you can alternatively be more specific about where you place the values. For example, you could insert short_array into array[2, 0:3, 0] directly before inserting the extra 0.
In the next section, you’ll learn a little about structured arrays and how they can be used to reconcile the differences between different arrays.
NumPy Example 2: Reconciling Data Using Structured Arrays
With the NumPy arrays you created in the previous section, there was no way of knowing the meaning of each column’s data. Wouldn’t it be nice if you could reference specific columns by meaningful names instead of index numbers? So, for example, instead of using student_grades = results[:, 1], you could instead use student_grades = results["exam_grade"]. Good news! You can do this by creating a structured array.
Creating a Structured Array
A structured array is a NumPy array with a data type composed of a set of tuples, each containing a field name and a regular data type. Once you’ve defined these, you can then access and modify each individual field using its field name.
The code below provides an example of how to create and reference a structured NumPy array:
1>>> import numpy as np
2
3>>> race_results = np.array(
4... [
5... ("At The Back", 1.2, 3),
6... ("Fast Eddie", 1.3, 1),
7... ("Almost There", 1.1, 2),
8... ],
9... dtype=[
10... ("horse_name", "U12"),
11... ("price", "f4"),
12... ("position", "i4"),
13... ],
14... )
15
16>>> race_results["horse_name"]
17array(['At The Back', 'Fast Eddie', 'Almost There'], dtype='<U12')
18
19>>> np.sort(race_results, order="position")[
20... ["horse_name", "price"]
21... ]
22array([('Fast Eddie', 1.3), ('Almost There', 1.1), ('At The Back', 1.2)],
23 dtype={'names': ['horse_name', 'price'],
24 ⮑ 'formats': ['<U12', '<f4'],
25 ⮑ 'offsets': [0, 48], 'itemsize': 56})
26
27>>> race_results[race_results["position"] == 1]["horse_name"]
28array(['Fast Eddie'], dtype='<U12')
The structured array is defined in lines 3 to 14, but begin by looking at lines 4 to 8, which define what looks like a normal NumPy array. It consists of three rows and three columns of data relating to a horse race. You can easily pick out the names of the horses from the data, but you may struggle a little to work out what the other two numbers mean.
This array is actually a structured array because of the definition of its data types in line 9. Each data type consists of a field name and an associated data type. The three fields are horse_name, price, and position. Their associated data types are defined using array interface protocol codes. U12 defines a 12-character string, while f4 and i4 specify 4-byte floating-point and integer formats, respectively.
Once you’ve set up your structured array, you can use these field names to reference columns. In line 16, you used "horse_name" to view an array of racehorse names. To find the finishing order, you passed the "position" field into the np.sort() function in line 19. This sorted the runners into their finishing order. You then filtered the output to show only horse_name and price. Finally, in line 27, you selected the name of the winning horse.
Reconciling Different Arrays
Including field names in your NumPy arrays has many useful purposes. Suppose you wanted to match records by matching field names in separate NumPy arrays. To do this, you join your arrays together so that only matching records from each array are shown. This idea will be familiar to you if you’ve ever performed an SQL inner join between two relational database tables. The term inner is used to define the join used here.
In this section, you’ll work with two new files: issued_checks.csv and cashed_checks.csv.
The issued_checks.csv file contains four fields: check_ID, Payee, Amount, and Date_Issued. This file simulates a set of checks issued by your business to its creditors:
issued_checks.csv
Check_ID,Payee,Amount,Date_Issued
1341,K Starmer,150.00,2024-03-29
1342,R Sunak,175.00,2024-03-29
1343,L Truss,30.00,2024-03-29
1344,B Johnson,45.00,2024-03-22
1345,T May,65.00,2024-03-22
1346,D Cameron,430.00,2024-03-22
1347,G Brown,100.00,2024-03-15
1348,T Blair,250.00,2024-03-15
1349,J Major,500.00,2024-03-15
1350,M Thatcher,220.00,2024-03-15
The cashed_checks.csv file contains only three fields: check_ID, Amount, and Date_Cashed. This file simulates a set of checks cashed by the creditors of your business:
cashed_checks.csv
Check_ID,Amount,Date_Cashed
1341,150.00,2024-04-12
1342,175.00,2024-04-16
1343,30.00,2024-04-12
1345,65.00,2024-04-12
1346,430.00,2024-04-08
1349,500.00,2024-04-08
1350,220.00,2024-04-15
Say that you wanted to see the payee, date_issued, and date_cashed for checks that were cashed. If you look carefully, then you’ll see that the Payee and Date_Issued details aren’t included in cashed_checks.csv, while the other two fields are. To see all the data you require, you’ll need to join both files together.
Add the issued_checks.csv and cashed_checks.csv files to your program’s folder, then run this code:
1>>> import numpy.lib.recfunctions as rfn
2>>> from pathlib import Path
3
4>>> issued_dtypes = [
5... ("id", "i8"),
6... ("payee", "U10"),
7... ("amount", "f8"),
8... ("date_issued", "U10"),
9... ]
10
11>>> cashed_dtypes = [
12... ("id", "i8"),
13... ("amount", "f8"),
14... ("date_cashed", "U10"),
15... ]
16
17>>> issued_checks = np.loadtxt(
18... Path("issued_checks.csv"),
19... delimiter=",",
20... dtype=issued_dtypes,
21... skiprows=1,
22... )
23
24>>> cashed_checks = np.loadtxt(
25... Path("cashed_checks.csv"),
26... delimiter=",",
27... dtype=cashed_dtypes,
28... skiprows=1,
29... )
30
31>>> cashed_check_details = rfn.rec_join(
32... "id",
33... issued_checks,
34... cashed_checks,
35... jointype="inner",
36... )
37
38>>> cashed_check_details[
39... ["payee", "date_issued", "date_cashed"]
40... ]
41array([('K Starmer', '2024-03-29', '2024-04-12'),
42 ('R Sunak', '2024-03-29', '2024-04-16'),
43 ('L Truss', '2024-03-29', '2024-04-12'),
44 ('T May', '2024-03-22', '2024-04-12'),
45 ('D Cameron', '2024-03-22', '2024-04-08'),
46 ('J Major', '2024-03-15', '2024-04-08'),
47 ('M Thatcher', '2024-03-15', '2024-04-15')],
48 dtype={'names': ['payee', 'date_issued', 'date_cashed'],
49 ⮑'formats': ['<U10', '<U10', '<U10'],
50 ⮑'offsets': [8, 64, 104], 'itemsize': 144})
To join together two NumPy arrays, you use one of several recarray helper functions, which allow you to work with structured arrays. To access these, you must first import the numpy.lib.recfunctions library module. Once again, you’ll use the pathlib library to access the files. They’re imported in lines 1 and 2.
In lines 4 to 15, you create two Python lists of tuples that define the data types to be used for both the issued_checks.csv file and the cashed_checks.csv file. These are fed into the dtype parameter of np.loadtext() to allow it to read the files into the NumPy structured arrays correctly.
The actual files are read by your code in lines 17 to 29. You use np.loadtext() as you did earlier, but this time you set its skiprows parameter to 1. This will ensure that the first row of each file is ignored because they each contain header information and not data.
The two NumPy arrays read in from the files are then joined together in lines 31 to 36 using the .rec_join() helper function. This function uses four parameters. The first parameter defines the field with which the two arrays will be joined. In this case, you want to join the arrays based on their id fields, which contain unique numbers identifying each check in both files.
Next, you pass in the names of the arrays to be joined as the second and third parameters.
The final parameter is the most interesting. By specifying jointype="inner", you perform an inner join. This means the resulting array will contain only matching records from both files, providing full details of all cashed checks. Any record whose id appears in one file but not the other will not appear in the output.
To confirm that the join has worked, look at the output produced by lines 38 to 40. The cashed_check_details array contains the payee, date_issued, and date_cashed data. The first two of these fields came from the issued_checks.csv file, while the latter came from cashed_checks.csv. As you can see, the records have been matched correctly.
Dealing With Duplicate Field Names
Now suppose you also want to view the check amount. You might be tempted to try this:
>>> cashed_check_details[
... [
... "payee",
... "date_issued",
... "date_cashed",
... "amount",
... ]
... ]
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
KeyError: 'amount'
The KeyError occurred because the amount field no longer exists in the joined array. This happened because both of the original data files contained an amount field. It’s not possible to have a structured array containing two fields with the same label. The join operation renamed them uniquely.
To find out what they’re now called, you first view the data types of the joined array. You access them through the .dtype property:
>>> cashed_check_details.dtype
dtype([('id', '<i8'), ('payee', '<U10'), ('amount1', '<f8'), ('amount2', '<f8'),
⮑ ('date_issued', '<U10'), ('date_cashed', '<U10')])
If you look carefully at the output, you’ll notice that the original amount fields have been renamed amount1 and amount2. To see their contents, you must use these names. In this scenario, both fields contain the same data, so it doesn’t matter which of them you choose:
>>> cashed_check_details[
... [
... "payee",
... "date_issued",
... "date_cashed",
... "amount1",
... ]
... ]
array([('K Starmer', '2024-03-29', '2024-04-12', 150.),
('R Sunak', '2024-03-29', '2024-04-16', 175.),
('L Truss', '2024-03-29', '2024-04-12', 30.),
('T May', '2024-03-22', '2024-04-12', 65.),
('D Cameron', '2024-03-22', '2024-04-08', 430.),
('J Major', '2024-03-15', '2024-04-08', 500.),
('M Thatcher', '2024-03-15', '2024-04-15', 220.)],
dtype={'names': ['payee', 'date_issued', 'date_cashed', 'amount1'],
⮑ 'formats': ['<U10', '<U10', '<U10', '<f8'],
⮑'offsets': [8, 64, 104, 48], 'itemsize': 144})
This time the check amounts are included in your output.
Now that you know what checks you’ve issued and what’s been cashed, you may want to see a list of checks that are currently uncashed:
1>>> outstanding_checks = [
2... check_id
3... for check_id in issued_checks["id"]
4... if check_id not in cashed_checks["id"]
5... ]
6
7>>> outstanding_checks
8[np.int64(1344), np.int64(1347), np.int64(1348)]
9
10>>> [int(check_id) for check_id in outstanding_checks]
11[1344, 1347, 1348]
In line 1, you create a list named outstanding_checks using a list comprehension. This will contain a list of checks with an id value that’s in the issued_checks array but not in the cashed_checks array. These are still outstanding.
Again, by using the field name, you can quickly specify the fields to use for the comparison and keep your code highly readable.
For convenience, you extract the numbers from your resulting array using a second list comprehension in line 10.
For completeness, you might want to find out if there have been any checks cashed fraudulently against your account that you haven’t issued:
>>> [
... check_id
... for check_id in cashed_checks["id"]
... if check_id not in issued_checks["id"]
... ]
[]
The empty list produced shows that, thankfully, there are none. Phew!
Dealing With Duplicate Key Values
Before joining NumPy arrays, it’s important to make sure there are no duplicate key values. These can cause undesirable results because they create one-to-many relationships. In cases where your duplicate keys are valid, for example, where you need to perform a one-to-many join, a better alternative is to use the merge() function in pandas.
The code below uses the check_list_duplicates.csv file. This file has the same structure as the issued_checks.csv file you used previously, but there’s a duplicate record:
1>>> from pathlib import Path
2>>> import numpy.lib.recfunctions as rfn
3
4>>> issued_dtypes = [
5... ("id", "i8"),
6... ("payee", "U10"),
7... ("amount", "f8"),
8... ("date_issued", "U10"),
9... ]
10
11>>> issued_checks = np.loadtxt(
12... Path("check_list_duplicates.csv"),
13... delimiter=",",
14... dtype=issued_dtypes,
15... skiprows=1,
16... )
17
18>>> rfn.find_duplicates(np.ma.asarray(issued_checks))
19masked_array(data=[(1344, 'B Johnson', 45.0, '2024-03-22'),
20 (1344, 'B Johnson', 45.0, '2024-03-22')],
21 mask=[(False, False, False, False),
22 (False, False, False, False)],
23 fill_value=(999999, 'N/A', 1e+20, 'N/A'),
24 dtype=[('id', '<i8'), ('payee', '<U10'),
25 ⮑ ('amount', '<f8'), ('date_issued', '<U10')])
To find duplicate rows in a structured array, you can use the .find_duplicates() helper function. This function requires that you pass it a masked array, which is an array that may have missing or invalid entries. Although your array is fine in this respect, you need to convert it to a masked array before passing it to the function. You can see from line 19 of the output that there’s a duplicate row—the 1344 record occurs twice.
To get rid of it, you can use the np.unique() function:
>>> issued_checks = np.unique(issued_checks, axis=0)
>>> issued_checks
array([(1341, 'K Starmer', 150., '2024-03-29'),
(1342, 'R Sunak', 175., '2024-03-29'),
(1343, 'L Truss', 30., '2024-03-29'),
(1344, 'B Johnson', 45., '2024-03-22'),
(1345, 'T May', 65., '2024-03-22'),
(1346, 'D Cameron', 430., '2024-03-22'),
(1347, 'G Brown', 100., '2024-03-15'),
(1348, 'T Blair', 250., '2024-03-15'),
(1349, 'J Major', 500., '2024-03-15'),
(1350, 'M Thatcher', 220., '2024-03-15')],
dtype=[('id', '<i8'), ('payee', '<U10'),
⮑ ('amount', '<f8'), ('date_issued', '<U10')])
To remove duplicate rows, you pass your issued_checks array, along with axis=0 to np.unique() to tell it to remove rows. This creates a fresh array and any duplicate rows are removed, leaving only one instance. If you look at the output, you’ll see the 1344 row now appears only once.
Testing Your Skills: Joining Arrays Together
It’s time for your next challenge. See if you can solve this exercise:
An airline holds data on its passengers in two separate files: passengers.csv and passports.csv file. Take a look at both files, which you’ll find in the downloadable materials, to familiarize yourself with their content. Now, use your skills to solve the following tasks:
Task 1: Produce a structured array containing the first name, last name, and nationality of each passenger.
Task 2: Determine if there any passengers on the list who don’t have passports.
Task 3: Determine if there any passports on the list that don’t belong to any passengers.
One possible solution to Task 1 is:
1>>> import numpy as np
2>>> import numpy.lib.recfunctions as rfn
3
4>>> passenger_dtype = [
5... ("passenger_no", "i8"),
6... ("first_name", "U20"),
7... ("last_name", "U20"),
8... ]
9
10>>> passport_dtype = [
11... ("passport_no", "i8"),
12... ("passenger_no", "i8"),
13... ("nationality", "U20"),
14... ]
15
16>>> passengers = np.unique(
17... np.loadtxt(
18... "passengers.csv",
19... delimiter=",",
20... dtype=passenger_dtype,
21... skiprows=1,
22... ),
23... axis=0,
24... )
25
26>>> passports = np.unique(
27... np.loadtxt(
28... "passports.csv",
29... delimiter=",",
30... dtype=passport_dtype,
31... skiprows=1,
32... ),
33... axis=0,
34... )
35
36>>> flight_details = rfn.rec_join(
37... "passenger_no",
38... passengers,
39... passports,
40... jointype="inner",
41... )
42
43>>> flight_details[
44... ["first_name", "last_name", "nationality"]
45... ]
46rec.array([('Olivia', 'Smith', 'British'), ('Amelia', 'Jones', 'British'),
47 ('Isla', 'Williams', 'American'),
48 ('Ava', 'Taylor', 'American'), ('Ivy', 'Brown', 'Austrian'),
49 ('Freya', 'Davies', 'Norwegian'), ('Lily', 'Evans', 'French'),
50 ('Florence', 'Wilson', 'German'), ('Mia', 'Thomas', 'Danish'),
51 ('Willow', 'Johnson', 'Dutch'), ('Noah', 'Roberts', 'Dutch'),
52 ('Oliver', 'Robinson', 'French'),
53 ('George', 'Thompson', 'Danish'),
54 ('Muhammad', 'Walker', 'Dutch'), ('Leo', 'White', 'British'),
55 ('Harry', 'Edwards', 'American'),
56 ('Oscar', 'Hughes', 'Spanish'),
57 ('Archie', 'Green', 'Norwegian'), ('Henry', 'Hall', 'British')],
58 dtype={'names': ['first_name', 'last_name', 'nationality'],
59 ⮑ 'formats': ['<U20', '<U20', '<U20'],
60 ⮑ 'offsets': [8, 88, 176], 'itemsize': 256})
You start off by creating two lists, passenger_dtype and passport_dtype, as shown in lines 4 and 10. These each store tuples that will be used to define the data types of each column in the structured arrays that will hold the file data. Each tuple consists of a field name and a data type. You may recall that i8 specifies an integer, while U20 defines a twenty-character string.
In lines 16 and 26, you read the data from each file into the passengers and passports arrays, respectively. You also use np.unique() to ensure duplicate records are removed. If you forget to do this, you’ll see some N/A values appear in your results. Notice that you’re skipping the top line of each file because it contains headings.
In line 36, you join both of your passengers and passports arrays together using their passenger_no fields as the join key. This is an inner join, meaning only passengers with passports are included in the output.
In line 43, you display the required flight details. The output shows the passengers’ names from the passengers.csv file and their nationalities from the passports.csv file. This approach takes advantage of the convenience and clarity of having field names to extract the data.
One possible solution to Task 2 is:
>>> passengers_without_passports = [
... passenger
... for passenger in passengers["passenger_no"]
... if passenger not in passports["passenger_no"]
... ]
>>> passengers_without_passports
[np.int64(14)]
To answer this question you use a list comprehension that loops through each passenger_no in your passengers array and returns only those that don’t appear in your passports array. In this case, passenger 14 hasn’t provided passport details.
One possible solution to Task 3 is:
>>> passports_without_passengers = [
... passenger
... for passenger in passports["passenger_no"]
... if passenger not in passengers["passenger_no"]
... ]
>>> passports_without_passengers
[np.int64(21)]
To answer this question you again use a list comprehension, but this time it loops through each passenger_no in your passports array and returns only those that don’t appear in your passengers array. In this case, the passport belonging to the non-existent passenger 21 is revealed.
In the next section, you’ll learn how to analyze hierarchical data.
NumPy Example 3: Analyzing and Charting Hierarchical Data
Hierarchical data is data that consists of different levels, with each level being linked to those immediately above and below. It’s often drawn using a tree diagram and different levels are often described as having a parent-child relationship.
For example, you may have an organization with several departments, with each department containing several staff members. There’s a hierarchical relationship between the organization, its departmental data, and the data of the staff members who work in each department.
Structured arrays are well-suited for joining hierarchical data because they allow you to reference data by labels. To use NumPy with hierarchical data, you can consolidate it into a single array.
Note: When joining NumPy arrays with rec_join(), you’re limited to creating inner joins. If you need more thorough join capabilities, then you should consider using the pandas merge() function instead.
In this section, you’ll look at analyzing a share portfolio. A share portfolio is the name given to the collection of shares held in a variety of companies. Creating a portfolio is a smart strategy for investors as it helps to spread investment risk. The idea is that losses incurred in some shares will be offset by gains in others.
Your share portfolio contains hierarchical data because it consists of multiple investments, each with its own collection of daily price movements. By analyzing this data, you can see how well your portfolio is performing.
Creating the Blank Array
The data used in this section simulates some hierarchical data. Suppose you maintain a file containing a list of the various companies you hold shares in. In this example, you’ll find this information in the portfolio.csv file included in your downloads. Here it is shown below:
portfolio.csv
Company,Sector
Company_A,technology
Company_B,finance
Company_C,healthcare
Company_D,technology
Company_E,finance
Company_F,healthcare
The Company column contains the company names, while the Sector column contains the sectors the company belongs to.
Each day, over the course of a week, you download the share prices for each of the companies you hold an interest in and add them to a structured NumPy array named portfolio. You’ll find each day’s price data in a series of separate files named share_prices-n.csv, where n is a number between one and five. For example, share_prices-1.csv contains the prices for Monday, share_prices-2.csv the prices for Tuesday, and so on.
A sample share prices file is shown below:
share_prices-1.csv
Company,mon
Company_A,100.5
Company_B,200.1
Company_C,50.3
Company_D,110.5
Company_E,200.1
Company_F,55.3
In this share_prices-1.csv file, there are two columns. The Company column displays the names of the companies, while the mon column shows the prices of each company share for Monday. The rest of the files follow a similar pattern, apart from their day columns, which are different.
To analyze this data, your portfolio array will need seven fields. In addition to the company name and the sector it belongs to, you’ll also need five fields to hold the daily prices for each company. The first two fields will be strings, while the rest will be floats.
The code shown below creates the initial portfolio array:
1>>> import numpy as np
2>>> from pathlib import Path
3
4>>> days = ["mon", "tue", "wed", "thu", "fri"]
5>>> days_dtype = [(day, "f8") for day in days]
6>>> company_dtype = [("company", "U20"), ("sector", "U20")]
7
8>>> portfolio_dtype = np.dtype(company_dtype + days_dtype)
9>>> portfolio = np.zeros((6,), dtype=portfolio_dtype)
10>>> portfolio
11array([('', '', 0., 0., 0., 0., 0.), ('', '', 0., 0., 0., 0., 0.),
12 ('', '', 0., 0., 0., 0., 0.), ('', '', 0., 0., 0., 0., 0.),
13 ('', '', 0., 0., 0., 0., 0.), ('', '', 0., 0., 0., 0., 0.)],
14 dtype=[('company', '<U20'), ('sector', '<U20'), ('mon', '<f8'),
15 ⮑ ('tue', '<f8'), ('wed', '<f8'), ('thu', '<f8'), ('fri', '<f8')])
As before, you’ll use both the numpy and pathlib libraries, so you import these in lines 1 and 2.
To define the field names and data types of each field in your portfolio array, you begin by creating the three lists shown in lines 4, 5, and 6. The days_dtype list contains a series of tuples, one for each value in the days array you created in line 4, with a data type of f8, which represents a floating-point number. The company_dtype list contains definitions for the portfolio.csv file’s company data.
To create the actual data type object that will define the portfolio array’s data types, you concatenate the company_dtype and days_dtype lists together. Then, you cast the result to a dtype object using the np.dtype() function as shown in line 8.
The dtype object is then fed into the np.zeros() function as its dtype parameter. You also configure your array with a shape of (6,) to provide a separate row for each share’s data. This produces an array containing empty strings in the first two fields and zeros in the rest, as shown in the output.
Populating the Array
Now that you’ve created an array that’s large enough to store all of the data you need, the next step is to start populating it. To start, you’ll add in the details of the companies stored in portfolio.csv:
1>>> companies = np.loadtxt(
2... Path("portfolio.csv"),
3... delimiter=",",
4... dtype=company_dtype,
5... skiprows=1,
6... ).reshape((6,))
7
8>>> portfolio[["company", "sector"]] = companies
9>>> portfolio
10array([('Company_A', 'technology', 0., 0., 0., 0., 0.),
11 ('Company_B', 'finance', 0., 0., 0., 0., 0.),
12 ('Company_C', 'healthcare', 0., 0., 0., 0., 0.),
13 ('Company_D', 'technology', 0., 0., 0., 0., 0.),
14 ('Company_E', 'finance', 0., 0., 0., 0., 0.),
15 ('Company_F', 'healthcare', 0., 0., 0., 0., 0.)],
16 dtype=[('company', '<U20'), ('sector', '<U20'), ('mon', '<f8'),
17 ⮑ ('tue', '<f8'), ('wed', '<f8'), ('thu', '<f8'), ('fri', '<f8')])
You once again use the loadtxt() function, this time to add data from portfolio.csv into a structured array named companies. Note that .reshape((6,)) was used in line 6 to give the array the same shape as the portfolio array you created earlier. This is necessary to insert companies into portfolio.
Line 8 is where the insertion takes place. The two companies array fields are inserted as the company and sector fields in portfolio, as you can see from the output.
All that remains to be done is the addition of the various daily share prices. The code for doing this is shown below:
1>>> share_prices_dtype = [("company", "U20"),("day", "f8"),]
2
3>>> for day, csv_file in zip(
4... days, sorted(Path.cwd().glob("share_prices-?.csv"))
5... ):
6... portfolio[day] = np.loadtxt(
7... csv_file.name,
8... delimiter=",",
9... dtype=share_prices_dtype,
10... skiprows=1,
11... )["day"]
12
13>>> portfolio
14array([('Company_A', 'technology', 100.5, 101.2, 102. , 101.8, 112.5),
15 ('Company_B', 'finance', 200.1, 199.8, 200.5, 201. , 200.8),
16 ('Company_C', 'healthcare', 50.3, 50.5, 51. , 50.8, 51.2),
17 ('Company_D', 'technology', 110.5, 101.2, 102. , 111.8, 97.5),
18 ('Company_E', 'finance', 200.1, 200.8, 200.5, 211. , 200.8),
19 ('Company_F', 'healthcare', 55.3, 50.5, 53. , 50.8, 52.2)],
20 dtype=[('company', '<U20'), ('sector', '<U20'), ('mon', '<f8'),
21 ⮑ ('tue', '<f8'), ('wed', '<f8'), ('thu', '<f8'), ('fri', '<f8')])
To begin, you create a list defining the two fields within each of the daily share_prices- files. To add this data into your main portfolio array, you once more create a loop that iterates over each of them but this time, you do so using Python’s built-in zip() function. The files will be processed in order as you learned about earlier.
In this case, you pass zip() the days list you defined earlier, as well as each of the files you’re processing. This produces a series of tuples, one for each day and file pair found. Each day in the tuple is assigned to the loop’s day variable, while each file is assigned to the csv_file variable.
Within the body of the loop each file is again read using np.loadtxt(), but this time, instead of keeping the entire file, only its day field data is stored. The first time around the loop, this data is inserted into the portfolio array’s mon field, the second time it gets inserted into the tue field, and so on. You did this by assigning the data read into portfolio[day] for each different day.
The final version of the array contains the company name as well as the sector to which it belongs. The final five numbers in each record are the Monday through Friday share prices in cents.
Now that you’ve combined your hierarchical data into a structured array, you can analyze it using the field names for simplicity. Suppose you want to extract a single company for a closer look:
>>> portfolio[portfolio["company"] == "Company_C"]
array([('Company_C', 'healthcare', 50.3, 50.5, 51., 50.8, 51.2)],
dtype=[('company', '<U20'), ('sector', '<U20'),
⮑ ('mon', '<f8'), ('tue', '<f8'), ('wed', '<f8'),
⮑ ('thu', '<f8'), ('fri', '<f8')])
Here, you extract a single company by locating it in the company column. To do this, you use portfolio["company"] == "Company_C", which selects those rows whose company column values match "Company_C". This method is far more intuitive than selecting rows with indexing.
Similarly, if you want to see how the technology companies in your portfolio perform on a Friday, you can also select those figures:
>>> portfolio[portfolio["sector"] == "technology"]["fri"]
array([112.5, 97.5])
To view the technology records, you use portfolio["sector"] == "technology". Then, to see only the Friday records, you filter them out using ["fri"].
Suppose you own 250 shares in each of your technology companies. You may want to see how much money they’re worth at the end of the week:
>>> portfolio[portfolio["sector"] == "technology"]["fri"] * 250 * 0.01
array([281.25, 243.75])
To do this, you use the techniques you’ve already learned to select the Friday figures for each of the technology parts of your portfolio. Then you multiply the figures by 250, which is the number of shares you own. Finally, you multiply the result by 0.01 to turn the amounts into dollars, keeping in mind that share prices are quoted in cents.
If you want the net worth, all you need to do is use sum():
>>> sum(portfolio[portfolio["sector"] == "technology"]["fri"] * 250 * 0.01)
np.float64(525.0)
The technology portion of your portfolio is worth $525.00.
As you can see, using structured arrays allows you to access data in a highly intuitive way. To take things further, you can also use this approach when using a structured array as the foundation for a Matplotlib chart. This is what you’ll do next.
Charting the Data
Suppose you want to display the analysis of the technology part of your portfolio on a chart. Again, because you’re working with a structured array, the code becomes intuitive:
1>>> import matplotlib.pyplot as plt
2
3>>> tech_mask = portfolio["sector"] == "technology"
4>>> tech_sector = portfolio[tech_mask]["company"]
5>>> tech_valuation = portfolio[tech_mask]["fri"] * 250 * 0.01
6
7>>> (
8... plt.bar(x=tech_sector, height=tech_valuation, data=tech_valuation)[0]
9... .set_color("g")
10... )
11>>> plt.xlabel("Tech Companies")
12>>> plt.ylabel("Friday Price ($)")
13>>> plt.title("Tech Share Valuation ($)")
14>>> plt.show()
You first create the tech_mask helper array. Next, you create two arrays that will be used in the plot. The tech_sector array, defined in line 4, contains the company names for each tech_sector company. The tech_valuation array, defined in line 5, contains the Friday valuations for each tech_sector company.
Lines 7 through 10 create the bar plot. The x-axis contains the tech_sector company names used to create the bars, while the height parameter contains their valuations. The tech_valuation values are what get plotted. Lines 11, 12, 13, and 14 produce labels for the axes and give the chart a title. Finally, the plot is displayed.
If you run the above code in a Jupyter Notebook, you don’t need to use plt.show(). If you run it in the standard Python REPL, object references will be shown after lines 11, 12 and 13. You can just ignore these, and they’ve been removed from the output for clarity.
The resulting plot is shown below:
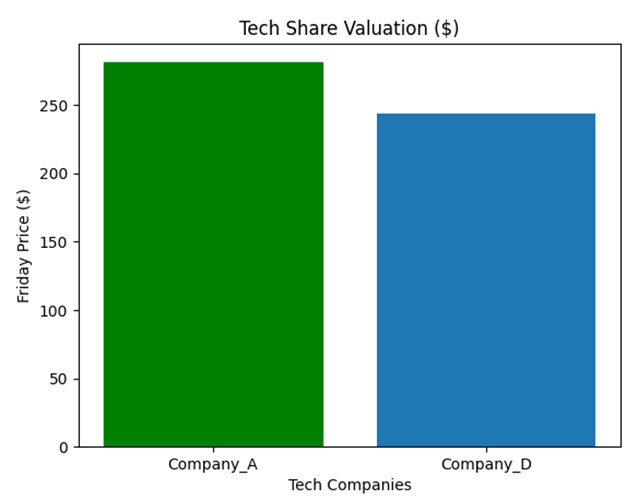
As you can see, Company_A appears to be doing better out of the two, albeit only slightly.
Testing Your Skills: Analyzing and Charting Hierarchical Data
Before you move on, here’s your third challenge:
You’ve been asked to collate the daily monthly average temperatures for each month of the year for analysis. The data is stored in the london_temperatures.csv, new_york_temperatures.csv, and rome_temperatures.csv files.
Using the skills you’ve learned, create a structured weather_data array with sensible field names and data types that contain four data values for each month. The first of these should contain the month, while the other three should contain the month’s temperature for each city.
Use your structured array to plot each monthly temperature for the three cities on a line plot.
One possible solution to creating the array is:
1>>> import numpy as np
2>>> from pathlib import Path
3
4>>> cities = ["london", "new_york", "rome"]
5>>> cities_dtype = [(city, "i8") for city in cities]
6>>> city_files_dtype = [("month", "U20"), ("temp", "i8")]
7>>> weather_data_dtype = np.dtype([("month", "U20")] + cities_dtype)
8
9>>> weather_data = np.zeros((12,), dtype=weather_data_dtype)
10>>> weather_data
11array([('', 0, 0, 0), ('', 0, 0, 0), ('', 0, 0, 0), ('', 0, 0, 0),
12 ('', 0, 0, 0), ('', 0, 0, 0), ('', 0, 0, 0), ('', 0, 0, 0),
13 ('', 0, 0, 0), ('', 0, 0, 0), ('', 0, 0, 0), ('', 0, 0, 0)],
14 dtype=[('month', '<U20'), ('london', '<i8'),
15 ⮑ ('new_york', '<i8'), ('rome', '<i8')])
16
17>>> for city in cities:
18... temps = np.loadtxt(
19... Path(f"{city}_temperatures.csv"),
20... delimiter=",",
21... dtype=city_files_dtype,
22... )
23... weather_data[["month", city]] = temps
24...
25
26>>> weather_data
27array([('Jan', 5, 2, 8), ('Feb', 7, 2, 9), ('Mar', 9, 4, 12),
28 ('Apr', 11, 11, 14), ('May', 14, 16, 21), ('Jun', 16, 22, 23),
29 ('Jul', 19, 25, 26), ('Aug', 19, 24, 24), ('Sep', 17, 20, 22),
30 ('Oct', 13, 14, 18), ('Nov', 10, 12, 13), ('Dec', 7, 9, 10)],
31 dtype=[('month', '<U20'), ('london', '<i8'),
32 ⮑ ('new_york', '<i8'), ('rome', '<i8')])
You start by creating a correctly sized array upfront to accept the data. As before, you use lists and list comprehensions to define each field and its data type before producing the weather_data array in lines 9 and 10. You can see the result in lines 11 to 14. Note that each month field is initialized with an empty string, while each integer field is set to 0.
In line 17, you start to loop over the individual cities. For London, you read the london_temperatures.csv file into a temps array, and in line 23 you assign its data to the month and london fields in your weather_data array. The New York and Rome data are read and added in the same manner. The month labels get overwritten for each city. But this is fine as long as they’re the same in all datasets.
Lines 27 through 32 show the complete array. One possibility for the line plot could be the following:
>>> import matplotlib.pyplot as plt
>>> plt.plot(weather_data["month"], weather_data["london"])
>>> plt.plot(weather_data["month"], weather_data["new_york"])
>>> plt.plot(weather_data["month"], weather_data["rome"])
>>> plt.ylabel("Temperature (C)")
>>> plt.xlabel("Month")
>>> plt.title("Average Monthly Temperatures")
>>> plt.legend(["London", "New York", "Rome"])
>>> plt.show()
To construct the plot, you plot the months along the x-axis and create a separate line for each of the three cities’ temperatures along the y-axis. You then add labels to both of your chart axes, and add a title and legend.
Your final output should look something like this:
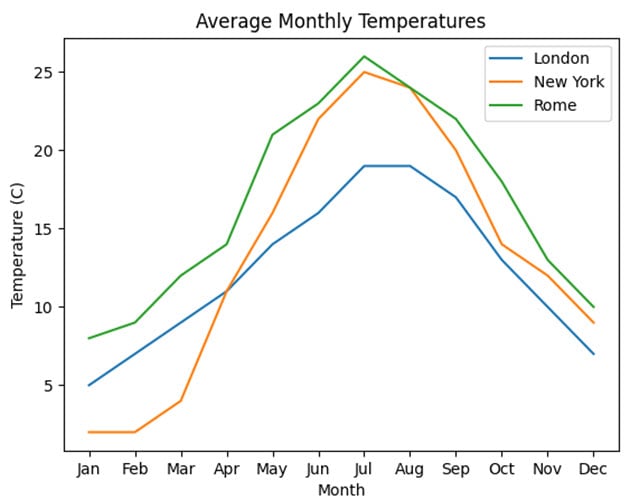
As you can see, of the three cities, Rome has consistently higher temperatures.
In the final section, you’ll learn about one of the main efficiencies of NumPy, and how to write functions that take advantage of it.
NumPy Example 4: Writing Your Own Vectorized Functions
One of the efficiencies of NumPy is its ability to perform calculations on entire arrays without the programmer having to write slow loops that manually loop through each row or element. Instead, NumPy uses the underlying C language to perform the calculation on the entire array. This is known as vectorization.
In this final section, you’ll work with the full_portfolio.csv file shown below:
full_portfolio.csv
Company,Sector,Mon,Tue,Wed,Thu,Fri
Company_A,technology,100.5,101.2,102,101.8,112.5
Company_B,finance,200.1,199.8,200.5,201.0,200.8
Company_C,healthcare,50.3,50.5,51.0,50.8,51.2
Company_D,technology,110.5,101.2,102,111.8,97.5
Company_E,finance,200.1,200.8,200.5,211.0,200.8
Company_F,healthcare,55.3,50.5,53.0,50.8,52.2
This data will look familiar to you since it’s a merger of the files used in the previous section. Each column’s heading has the same meaning as before.
The code below shows vectorization in action:
1>>> import numpy as np
2>>> from pathlib import Path
3
4>>> share_dtypes = [
5... ("company", "U20"),
6... ("sector", "U20"),
7... ("mon", "f8"),
8... ("tue", "f8"),
9... ("wed", "f8"),
10... ("thu", "f8"),
11... ("fri", "f8"),
12... ]
13
14>>> portfolio = np.loadtxt(
15... Path("full_portfolio.csv"),
16... delimiter=",",
17... dtype=share_dtypes,
18... skiprows=1,
19... )
20
21>>> portfolio["fri"] - portfolio["mon"]
22array([ 12. , 0.7, 0.9, -13. , 0.7, -3.1])
After constructing the portfolio structured array, you decide to see how your shares have performed over the week. To do this, you pick out two arrays—one containing the Monday share prices and one containing those from Friday. To view the weekly change, you subtract the Monday prices from the Friday prices.
Notice in line 21, that although you’re subtracting one array from the other, NumPy subtracts each individual element of the arrays without you having to write code that loops through them individually. This is vectorization.
Now suppose that you get an extra 10% bonus on shares that have risen more than 1% in value during the week you’re analyzing. To find your profit including the bonus, you need to consider two cases—those shares getting the bonus and those that don’t. To do this, you might try the following:
1>>> def profit_with_bonus(first_day, last_day):
2... if last_day >= first_day * 1.01:
3... return (last_day - first_day) * 1.1
4... else:
5... return last_day - first_day
6...
7>>> profit_with_bonus(portfolio["mon"], portfolio["fri"])
8Traceback (most recent call last):
9 ...
10ValueError: The truth value of an array with more than one element is ambiguous.
11⮑ Use a.any() or a.all()
In lines 1 through 5, you’ve defined a function named profit_with_bonus() that will return your profit based on how your share’s price has risen over the week. If the profit is more than 1%, then you add another 10% as a bonus.
When you call your function in line 7, it raises a ValueError exception because it can’t interpret the arrays you’ve passed to it. It’ll only work if you pass it two numbers. To make this function work with arrays, read on.
Adding Vectorization Functionality With np.vectorize()
To make your profit_with_bonus() function work with arrays, you’ll need to turn it into a vectorized function. One way to do this is to use the np.vectorize() function. This function will return a version of your original function, but one that takes arrays as input instead of scalars.
The code shown below transforms your function into a vectorized function:
1>>> def profit_with_bonus(first_day, last_day):
2... if last_day >= first_day * 1.01:
3... return (last_day - first_day) * 1.1
4... else:
5... return last_day - first_day
6...
7>>> vectorized_profit_with_bonus = np.vectorize(profit_with_bonus)
8>>> vectorized_profit_with_bonus(portfolio["mon"], portfolio["fri"])
9array([ 13.2 , 0.7 , 0.99, -13. , 0.7 , -3.1 ])
To turn your profit_with_bonus() function into a vectorized version, you pass it into the np.vectorize() function in line 7. The vectorized version is then assigned to the vectorized_profit_with_bonus name.
In line 8, you call the new function and pass to it the arrays you want it to analyze. In this example, the output shows your profit, including the added bonus. If you compare the numbers to the straightforward profit calculation you did earlier, then you’ll notice that you’ve gotten a bonus for your shares in the first and third company. Notice that you didn’t need to change the original function in lines 1 to 5.
One final point about using np.vectorize() for you to note is that the original function is still available should you need it:
>>> in_profit(3, 5)
2.2
This time, a single value is returned. Since 5 - 3 is 2, and 2 is more than 1% of 3, you add 10% to get 2.2.
Adding Vectorization Functionality With @np.vectorize
As you’ve just seen, the np.vectorize() function creates a second function, meaning the original version is still available if you need to use it. As an alternative, you could use np.vectorize as a decorator instead:
1>>> @np.vectorize
2... def profit_with_bonus(first_day, last_day):
3... if last_day >= first_day * 1.01:
4... return (last_day - first_day) * 1.1
5... else:
6... return last_day - first_day
7...
8>>> profit_with_bonus(portfolio["mon"], portfolio["fri"])
9array([ 13.2 , 0.7 , 0.99, -13. , 0.7 , -3.1 ])
By applying @np.vectorize to your function in line 1, the profit_with_bonus() function is transformed into a vectorized version. This new version works the same way as the vectorized_profit_with_bonus() function you saw earlier. To use it, call profit_with_bonus() as you normally would, but make sure to pass your arrays into it. This will return the same profit array as before.
Unlike the version created by the np.vectorize() function, the original scalar version of profit_with_bonus() no longer exists. However, if you pass it two scalars, it’ll still work but return the result as an array:
>>> in_profit(3, 5)
array(2.2)
As you can see, the output is an array this time.
Using Existing Vectorization Functionality With np.where()
Many of the available NumPy functions already support vectorization. It’s a good idea to consult the documentation to see if there’s already a function available to suit your needs. To find your profit from the shares, for example, you could have used the np.where() function to account for both the bonus case and the regular profit case instead of writing your own function:
>>> np.where(
... portfolio["fri"] > portfolio["mon"] * 1.01,
... (portfolio["fri"] - portfolio["mon"]) * 1.1,
... portfolio["fri"] - portfolio["mon"],
... )
array([ 13.2 , 0.7 , 0.99, -13. , 0.7 , -3.1 ])
This time, you pass your condition into np.where(). Unlike your original profit_with_bonus() function, .where() supports vectorization natively. In this case, you use .where() by passing it a comparison between two arrays. You also pass it two calculated arrays. The first represents the profit with a 10% added bonus, while the second is the plain profit.
Now, np.where() will evaluate the condition for each element, and choose the corresponding element from one of the two arrays. If the condition is True, then it uses the element from the first array, and if it’s False, it picks the element from the second one.
Testing Your Skills: Writing a Vectorized Function
It’s time to dive into your final exercise challenge. You’re almost finished.
You’ve been asked to create two arrays containing the minimum and maximum temperatures from the weather_data array you created in your last challenge. If you’ve not already done so, construct the weather_data array by running the first block of code in the solution to the Analyzing and Charting Hierarchical Data exercise before going any further.
Then, write a function named find_min_max() that accepts three scalar values, for example, integers, and returns the maximum and minimum values. Now, use either @np.vectorize or np.vectorize() to enhance your function to work with NumPy arrays. Make sure you can call the vectorized version of your function by its original name.
As a starting point, your initial function could look something like this:
>>> def find_min_max(first, second, third):
... min_temp = min(first, second, third)
... max_temp = max(first, second, third)
... return min_temp, max_temp
>>> find_min_max(2, 1, 3)
(1, 3)
Your initial find_min_max() function takes three scalar values and returns the lowest and highest values. If you try it using data from your weather_data array, it won’t work:
>>> london_temps = weather_data["london"]
>>> new_york_temps = weather_data["new_york"]
>>> rome_temps = weather_data["rome"]
>>> find_min_max(london_temps, new_york_temps, rome_temps)
>>> # ...
ValueError: The truth value of an array with more than one element is ambiguous.
As you can see, you’ve raised a ValueError exception. You’ll need to update it to work with arrays.
A possible solution using @np.vectorize could be:
>>> @np.vectorize
... def find_min_max(first, second, third):
... min_temp = min(first, second, third)
... max_temp = max(first, second, third)
... return min_temp, max_temp
>>> london_temps = weather_data["london"]
>>> new_york_temps = weather_data["new_york"]
>>> rome_temps = weather_data["rome"]
>>> find_min_max(london_temps, new_york_temps, rome_temps)
(array([ 2, 2, 4, 11, 14, 16, 19, 19, 17, 13, 10, 7]),
⮑ array([ 8, 9, 12, 14, 21, 23, 26, 24, 22, 18, 13, 10]))
To use the vectorized version of your find_min_max() function, you decorate it with @np.vectorize. To call the vectorized function, you pass in the three arrays of weather data directly. The decorator allows it to understand arrays as inputs and produce them as outputs. The function uses the built-in min() and max() functions to analyze each of the three elements for each month and returns the result in two arrays.
An alternative solution could be to revert back to your original function and use np.vectorize() instead:
>>> def find_min_max(first, second, third):
... min_temp = min(first, second, third)
... max_temp = max(first, second, third)
... return min_temp, max_temp
>>> london_temps = weather_data["london"]
>>> new_york_temps = weather_data["new_york"]
>>> rome_temps = weather_data["rome"]
>>> find_min_max = np.vectorize(find_min_max)
>>> find_min_max(london_temps, new_york_temps, rome_temps)
(array([ 2, 2, 4, 11, 14, 16, 19, 19, 17, 13, 10, 7]),
⮑ array([ 8, 9, 12, 14, 21, 23, 26, 24, 22, 18, 13, 10]))
To use the vectorized version of your find_min_max() function, you pass it into np.vectorize(), and by assigning the output back to the variable find_min_max, you can then call the vectorized version using its original name. To call this vectorized version, you pass in the three arrays of weather data directly.
Conclusion
You’ve now gained insight into some interesting use cases for NumPy. Although NumPy is essentially a Python library that allows you to work with multidimensional array objects, you now have a better understanding of how its features can be applied in several different scenarios.
In this tutorial, you’ve learned how to:
- Construct NumPy array objects from external data files
- Find matching data between different NumPy arrays
- Extract named data and construct basic charts
- Transform your own function into a vectorized function
While this tutorial isn’t exhaustive, hopefully it’s given you some helpful ideas for using NumPy. If you want to learn more about other aspects of NumPy, you can work through some of our other NumPy tutorials. Then you’ll be more than ready to read through the NumPy documentation and apply it to any problems you may need to solve.
To complement your existing skills, you might consider broadening your knowledge by learning more about related libraries such as pandas or Polars. And don’t forget about Matplotlib!
Get Your Code: Click here to download the free sample code that you’ll use to work through NumPy practical examples.
Take the Quiz: Test your knowledge with our interactive “NumPy Practical Examples: Useful Techniques” quiz. You’ll receive a score upon completion to help you track your learning progress:
Interactive Quiz
NumPy Practical Examples: Useful TechniquesThis quiz will test your understanding of working with NumPy arrays. You won't find all the answers in the tutorial, so you'll need to do some extra investigating. By finding all the answers, you're sure to learn some interesting things along the way.
Watch Now This tutorial has a related video course created by the Real Python team. Watch it together with the written tutorial to deepen your understanding: NumPy Techniques and Practical Examples






