With train_test_split() from scikit-learn, you can efficiently divide your dataset into training and testing subsets to ensure unbiased model evaluation in machine learning. This process helps prevent overfitting and underfitting by keeping the test data separate from the training data, allowing you to assess the model’s predictive performance accurately.
By the end of this tutorial, you’ll understand that:
train_test_split()is a function insklearnthat divides datasets into training and testing subsets.x_trainandy_trainrepresent the inputs and outputs of the training data subset, respectively, whilex_testandy_testrepresent the input and output of the testing data subset.- By specifying
test_size=0.2, you use 20% of the dataset for testing, leaving 80% for training. train_test_split()can handle imbalanced datasets using thestratifyparameter to maintain class distribution.
You’ll learn how to use train_test_split() and apply these concepts in real-world scenarios, ensuring your machine learning models are evaluated with precision and fairness. In addition, you’ll explore related tools from sklearn.model_selection for further insights.
Get Your Code: Click here to download the free sample code that you’ll use to learn about splitting your dataset with scikit-learn’s train_test_split().
Take the Quiz: Test your knowledge with our interactive “Split Your Dataset With scikit-learn's train_test_split()” quiz. You’ll receive a score upon completion to help you track your learning progress:
Interactive Quiz
Split Your Dataset With scikit-learn's train_test_split()In this quiz, you'll test your understanding of how to use the train_test_split() function from the scikit-learn library to split your dataset into subsets for unbiased evaluation in machine learning.
The Importance of Data Splitting
Supervised machine learning is about creating models that precisely map the given inputs to the given outputs. Inputs are also called independent variables or predictors, while outputs may be referred to as dependent variables or responses.
How you measure the precision of your model depends on the type of a problem you’re trying to solve. In regression analysis, you typically use the coefficient of determination, root mean square error, mean absolute error, or similar quantities. For classification problems, you often apply accuracy, precision, recall, F1 score, and related indicators.
The acceptable numeric values that measure precision vary from field to field. You can find detailed explanations from Statistics By Jim, Quora, and many other resources.
What’s most important to understand is that you usually need unbiased evaluation to properly use these measures, assess the predictive performance of your model, and validate the model.
This means that you can’t evaluate the predictive performance of a model with the same data you used for training. You need evaluate the model with fresh data that hasn’t been seen by the model before. You can accomplish that by splitting your dataset before you use it.
Training, Validation, and Test Sets
Splitting your dataset is essential for an unbiased evaluation of prediction performance. In most cases, it’s enough to split your dataset randomly into three subsets:
-
The training set is applied to train or fit your model. For example, you use the training set to find the optimal weights, or coefficients, for linear regression, logistic regression, or neural networks.
-
The validation set is used for unbiased model evaluation during hyperparameter tuning. For example, when you want to find the optimal number of neurons in a neural network or the best kernel for a support vector machine, you experiment with different values. For each considered setting of hyperparameters, you fit the model with the training set and assess its performance with the validation set.
-
The test set is needed for an unbiased evaluation of the final model. You shouldn’t use it for fitting or validation.
In less complex cases, when you don’t have to tune hyperparameters, it’s okay to work with only the training and test sets.
Underfitting and Overfitting
Splitting a dataset might also be important for detecting if your model suffers from one of two very common problems, called underfitting and overfitting:
-
Underfitting is usually the consequence of a model being unable to encapsulate the relations among data. For example, this can happen when trying to represent nonlinear relations with a linear model. Underfitted models will likely have poor performance with both training and test sets.
-
Overfitting usually takes place when a model has an excessively complex structure and learns both the existing relations among data and noise. Such models often have bad generalization capabilities. Although they work well with training data, they usually yield poor performance with unseen test data.
You can find a more detailed explanation of underfitting and overfitting in Linear Regression in Python.
Prerequisites for Using train_test_split()
Now that you understand the need to split a dataset in order to perform unbiased model evaluation and identify underfitting or overfitting, you’re ready to learn how to split your own datasets.
You’ll use version 1.5.0 of scikit-learn, or sklearn. It has many packages for data science and machine learning, but for this tutorial, you’ll focus on the model_selection package, specifically on the function train_test_split().
Note: While this tutorial is tested with this specific version of scikit-learn, the features that you’ll use are core to the library and should work equivalently in other versions of scikit-learn as well.
You can install sklearn with pip:
$ python -m pip install "scikit-learn==1.5.0"
If you use Anaconda, then you probably already have it installed. However, if you want to use a fresh environment, ensure that you have the specified version or use Miniconda. Then you can install sklearn from Anaconda Cloud with conda install:
$ conda install -c anaconda scikit-learn=1.5.0
You’ll also need NumPy, but you don’t have to install it separately. You should get it along with sklearn if you don’t already have it installed. If you want to, you can refresh your NumPy knowledge and check out NumPy Tutorial: Your First Steps Into Data Science in Python.
Application of train_test_split()
You need to import train_test_split() and NumPy before you can use them. You can work in a Jupyter notebook or start a new Python REPL session, then you can start with the import statements:
>>> import numpy as np
>>> from sklearn.model_selection import train_test_split
Now that you have both imported, you can use them to split data into training sets and test sets. You’ll split inputs and outputs at the same time, with a single function call.
With train_test_split(), you only need to provide the arrays that you want to split. Additionally, you can also provide some optional arguments. The function usually returns a list of NumPy arrays but can also return a couple of other iterable types, such as SciPy sparse matrices, if appropriate:
sklearn.model_selection.train_test_split(*arrays, **options) -> list
The arrays parameter in the function signature of train_test_split() refers to the sequence of lists, NumPy arrays, pandas DataFrames, or similar array-like objects that hold the data that you want to split. All these objects together make up the dataset and must be of the same length.
In supervised machine learning applications, you’ll typically work with two such arrays:
- A two-dimensional array with the inputs (
x) - A one-dimensional array with the outputs (
y)
The options parameter indicates that you can customize the function’s behavior with optional keyword arguments:
-
train_sizeis the number that defines the size of the training set. If you provide afloat, then it must be between0.0and1.0and it will define the share of the dataset used for testing. If you provide anint, then it will represent the total number of the training samples. The default value isNone. -
test_sizeis the number that defines the size of the test set. It’s very similar totrain_size. You should provide eithertrain_sizeortest_size. If neither is given, then the default share of the dataset that will be used for testing is0.25, or 25 percent. -
random_stateis the object that controls randomization during splitting. It can be either anintor an instance ofRandomState. Setting the random state is useful if you need reproducibility. The default value isNone. -
shuffleis the Boolean object that determines whether to shuffle the dataset before applying the split. The default value isTrue. -
stratifyis an array-like object that, if notNone, determines how to use a stratified split.
Now it’s time to try data splitting! You’ll start by creating a simple dataset to work with. The dataset will contain the inputs in the two-dimensional array x and outputs in the one-dimensional array y:
>>> x = np.arange(1, 25).reshape(12, 2)
>>> y = np.array([0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 1, 0])
>>> x
array([[ 1, 2],
[ 3, 4],
[ 5, 6],
[ 7, 8],
[ 9, 10],
[11, 12],
[13, 14],
[15, 16],
[17, 18],
[19, 20],
[21, 22],
[23, 24]])
>>> y
array([0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 1, 0])
To get your data, you use arange(), which is very convenient for generating arrays based on numerical ranges. You also use .reshape() to modify the shape of the array returned by arange() and get a two-dimensional data structure.
You can split both input and output datasets with a single function call:
>>> x_train, x_test, y_train, y_test = train_test_split(x, y)
>>> x_train
array([[15, 16],
[21, 22],
[11, 12],
[17, 18],
[13, 14],
[ 9, 10],
[ 1, 2],
[ 3, 4],
[19, 20]])
>>> x_test
array([[ 5, 6],
[ 7, 8],
[23, 24]])
>>> y_train
array([1, 1, 0, 1, 0, 1, 0, 1, 0])
>>> y_test
array([1, 0, 0])
Given two arrays, like x and y here, train_test_split() performs the split and returns four arrays (in this case NumPy arrays) in this order:
x_train: The training part of the first array (x)x_test: The test part of the first array (x)y_train: The training part of the second array (y)y_test: The test part of the second array (y)
You probably got different results from what you see here. This is because dataset splitting is random by default. The result differs each time you run the function. However, this often isn’t what you want.
Sometimes, to make your tests reproducible, you need a random split with the same output for each function call. You can do that with the parameter random_state. The value of random_state isn’t important—it can be any non-negative integer. You could use an instance of numpy.random.RandomState instead, but that’s a more complex approach.
In the previous example, you used a dataset with twelve rows, or observations, and got a training sample with nine rows and a test sample with three rows. That’s because you didn’t specify the desired size of the training and test sets. By default, 25 percent of samples are assigned to the test set. This ratio is generally fine for many applications, but it’s not always what you need.
Typically, you’ll want to define the size of the test or training set explicitly, and sometimes you’ll even want to experiment with different values. You can do that with the parameters train_size or test_size.
Modify the code so you can choose the size of the test set and get a reproducible result:
>>> x_train, x_test, y_train, y_test = train_test_split(
... x, y, test_size=4, random_state=4
... )
>>> x_train
array([[17, 18],
[ 5, 6],
[23, 24],
[ 1, 2],
[ 3, 4],
[11, 12],
[15, 16],
[21, 22]])
>>> x_test
array([[ 7, 8],
[ 9, 10],
[13, 14],
[19, 20]])
>>> y_train
array([1, 1, 0, 0, 1, 0, 1, 1])
>>> y_test
array([0, 1, 0, 0])
With this change, you get a different result from before. Earlier, you had a training set with nine items and a test set with three items. Now, thanks to the argument test_size=4, the training set has eight items and the test set has four items. You’d get the same result with test_size=0.33 because 33 percent of twelve is approximately four.
There’s one more very important difference between the last two examples: You now get the same result each time you run the function. This is because you’ve fixed the random number generator with random_state=4.
The figure below shows what’s going on when you call train_test_split():
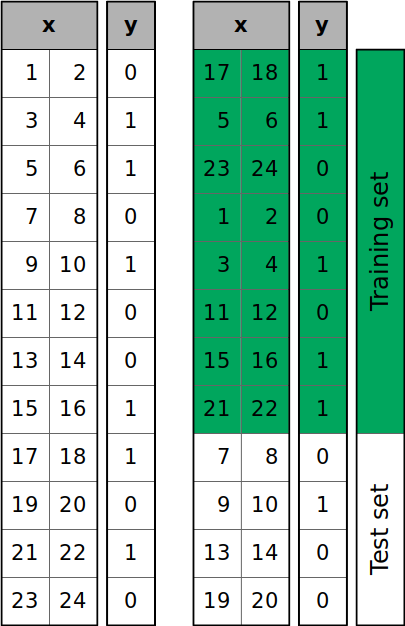
The samples of the dataset are shuffled randomly and then split into the training and test sets according to the size you defined.
You can see that y has six zeros and six ones. However, the test set has three zeros out of four items. If you want to (approximately) keep the proportion of y values through the training and test sets, then pass stratify=y. This will enable stratified splitting:
>>> x_train, x_test, y_train, y_test = train_test_split(
... x, y, test_size=0.33, random_state=4, stratify=y
... )
>>> x_train
array([[21, 22],
[ 1, 2],
[15, 16],
[13, 14],
[17, 18],
[19, 20],
[23, 24],
[ 3, 4]])
>>> x_test
array([[11, 12],
[ 7, 8],
[ 5, 6],
[ 9, 10]])
>>> y_train
array([1, 0, 1, 0, 1, 0, 0, 1])
>>> y_test
array([0, 0, 1, 1])
Now y_train and y_test have the same ratio of zeros and ones as the original y array.
Stratified splits are desirable in some cases, like when you’re classifying an imbalanced dataset, which is a dataset with a significant difference in the number of samples that belong to distinct classes.
Finally, you can turn off data shuffling and random split with shuffle=False:
>>> x_train, x_test, y_train, y_test = train_test_split(
... x, y, test_size=0.33, shuffle=False
... )
>>> x_train
array([[ 1, 2],
[ 3, 4],
[ 5, 6],
[ 7, 8],
[ 9, 10],
[11, 12],
[13, 14],
[15, 16]])
>>> x_test
array([[17, 18],
[19, 20],
[21, 22],
[23, 24]])
>>> y_train
array([0, 1, 1, 0, 1, 0, 0, 1])
>>> y_test
array([1, 0, 1, 0])
Now you have a split in which the first two-thirds of samples in the original x and y arrays are assigned to the training set and the last third to the test set. No shuffling. No randomness.
Supervised Machine Learning With train_test_split()
Now it’s time to see train_test_split() in action when solving supervised learning problems. You’ll start with a small regression problem that can be solved with linear regression before looking at a bigger problem. You’ll also see that you can use train_test_split() for classification as well.
Minimalist Example of Linear Regression
In this example, you’ll apply what you’ve learned so far to solve a small regression problem. You’ll learn how to create datasets, split them into training and test subsets, and use them for linear regression.
As always, you’ll start by importing the necessary packages, functions, or classes. You’ll need NumPy, LinearRegression, and train_test_split():
>>> import numpy as np
>>> from sklearn.linear_model import LinearRegression
>>> from sklearn.model_selection import train_test_split
Now that you’ve imported everything you need, you can create two small arrays, x and y, to represent the observations and then split them into training and test sets just as you did before:
>>> x = np.arange(20).reshape(-1, 1)
>>> y = np.array([5, 12, 11, 19, 30, 29, 23, 40, 51, 54, 74,
... 62, 68, 73, 89, 84, 89, 101, 99, 106])
>>> x
array([[ 0],
[ 1],
[ 2],
[ 3],
[ 4],
[ 5],
[ 6],
[ 7],
[ 8],
[ 9],
[10],
[11],
[12],
[13],
[14],
[15],
[16],
[17],
[18],
[19]])
>>> y
array([ 5, 12, 11, 19, 30, 29, 23, 40, 51, 54, 74, 62, 68,
73, 89, 84, 89, 101, 99, 106])
>>> x_train, x_test, y_train, y_test = train_test_split(
... x, y, test_size=8, random_state=0
... )
Your dataset has twenty observations, or x-y pairs. You specify the argument test_size=8, so the dataset is divided into a training set with twelve observations and a test set with eight observations.
Now you can use the training set to fit the model:
>>> model = LinearRegression().fit(x_train, y_train)
>>> model.intercept_
np.float64(3.1617195496417523)
>>> model.coef_
array([5.53121801])
LinearRegression creates the object that represents the model, while .fit() trains, or fits, the model and returns it. With linear regression, fitting the model means determining the best intercept (model.intercept_) and slope (model.coef_) values of the regression line.
Although you can use x_train and y_train to check the goodness of fit, this isn’t a best practice. An unbiased estimation of the predictive performance of your model is based on test data:
>>> model.score(x_train, y_train)
0.9868175024574795
>>> model.score(x_test, y_test)
0.9465896927715023
.score() returns the coefficient of determination, or R², for the data passed. Its maximum is 1. The higher the R² value, the better the fit. In this case, the training data yields a slightly higher coefficient. However, the R² calculated with test data is an unbiased measure of your model’s prediction performance.
This is how it looks on a graph:
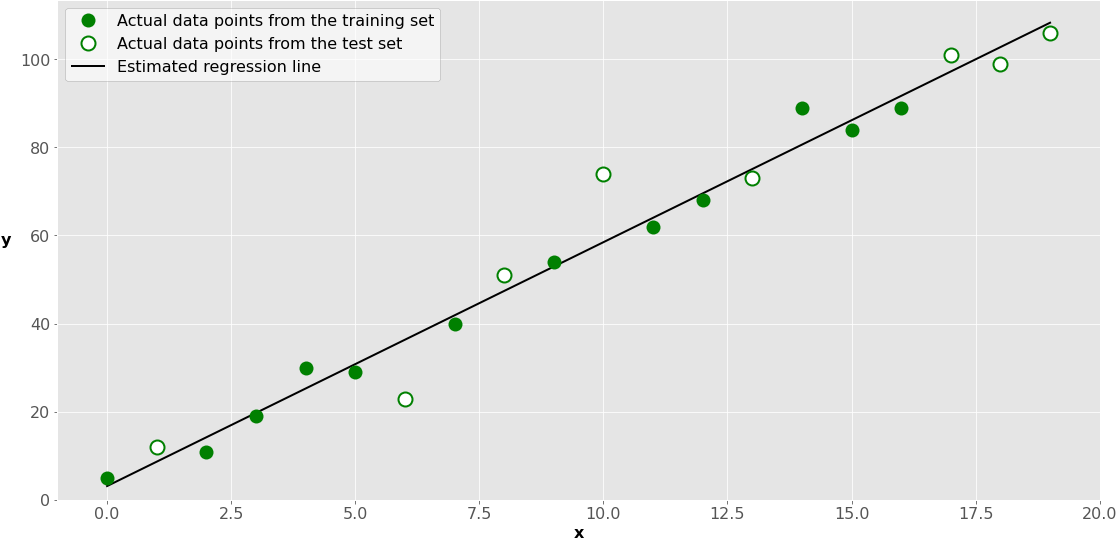
The green dots represent the x-y pairs used for training. The black line, called the estimated regression line, is defined by the results of model fitting: the intercept and the slope. So, it reflects the positions of the green dots only.
The white dots represent the test set. You use them to estimate the performance of the model (regression line) with data not used for training.
Regression Example
Now you’re ready to split a larger dataset to solve a regression problem. You’ll use the California Housing dataset, which is included in sklearn. This dataset has 20640 samples, eight input variables, and the house values as the output. You can retrieve it with sklearn.datasets.fetch_california_housing().
First, import train_test_split() and fetch_california_housing():
>>> from sklearn.datasets import fetch_california_housing
>>> from sklearn.model_selection import train_test_split
Now that you have both functions imported, you can get the data you’ll work with:
>>> x, y = fetch_california_housing(return_X_y=True)
As you can see, fetch_california_housing() with the argument return_X_y=True returns a tuple with two NumPy arrays:
- A two-dimensional array with the inputs
- A one-dimensional array with the outputs
The next step is to split the data the same way as before:
>>> x_train, x_test, y_train, y_test = train_test_split(
... x, y, test_size=0.4, random_state=0
... )
Now you have the training and test sets. The training data is contained in x_train and y_train, while the data for testing is in x_test and y_test.
When you work with larger datasets, it’s usually more convenient to pass the training or test size as a ratio. test_size=0.4 means that approximately 40 percent of samples will be assigned to the test data, and the remaining 60 percent will be assigned to the training data.
Finally, you can use the training set (x_train and y_train) to fit the model and the test set (x_test and y_test) for an unbiased evaluation of the model. In this example, you’ll apply three well-known regression algorithms to create models that fit your data:
- Linear regression with
LinearRegression() - Gradient boosting with
GradientBoostingRegressor() - Random forest with
RandomForestRegressor()
The process is pretty much the same as with the previous example:
- Import the classes you need.
- Create model instances using these classes.
- Fit the model instances with
.fit()using the training set. - Evaluate the model with
.score()using the test set.
Here’s the code that follows the steps described above for all three regression algorithms:
>>> from sklearn.linear_model import LinearRegression
>>> model = LinearRegression().fit(x_train, y_train)
>>> model.score(x_train, y_train)
0.6105322745695656
>>> model.score(x_test, y_test)
0.5982535501446862
>>> from sklearn.ensemble import GradientBoostingRegressor
>>> model = GradientBoostingRegressor(random_state=0).fit(x_train, y_train)
>>> model.score(x_train, y_train)
0.8083859166342285
>>> model.score(x_test, y_test)
0.7802104901623703
>>> from sklearn.ensemble import RandomForestRegressor
>>> model = RandomForestRegressor(random_state=0).fit(x_train, y_train)
>>> model.score(x_train, y_train)
0.9727449572570027
>>> model.score(x_test, y_test)
0.7933138227558006
You’ve used your training and test datasets to fit three models and evaluate their performance. The measure of accuracy obtained with .score() is the coefficient of determination. It can be calculated with either the training or test set. However, as you already learned, the score obtained with the test set represents an unbiased estimation of performance.
As mentioned in the documentation, you can provide optional arguments to LinearRegression(), GradientBoostingRegressor(), and RandomForestRegressor(). GradientBoostingRegressor() and RandomForestRegressor() use the random_state parameter for the same reason that train_test_split() does: to deal with randomness in the algorithms and ensure reproducibility.
For some methods, you may also need feature scaling. In such cases, you should fit the scalers with training data and use them to transform test data.
Classification Example
You can use train_test_split() to solve classification problems the same way you do for regression analysis. In machine learning, classification problems involve training a model to apply labels to, or classify, the input values and sort your dataset into categories.
In the tutorial Logistic Regression in Python, you’ll find an example of a handwriting recognition task. The example provides another demonstration of splitting data into training and test sets to avoid bias in the evaluation process.
Other Validation Functionalities
The package sklearn.model_selection offers a lot of functionalities related to model selection and validation, including the following:
- Cross-validation
- Learning curves
- Hyperparameter tuning
Cross-validation is a set of techniques that combine the measures of prediction performance to get more accurate model estimations.
One of the widely used cross-validation methods is k-fold cross-validation. In it, you divide your dataset into k (often five or ten) subsets, or folds, of equal size and then perform the training and test procedures k times. Each time, you use a different fold as the test set and all the remaining folds as the training set. This provides k measures of predictive performance, and you can then analyze their mean and standard deviation.
You can implement cross-validation with KFold, StratifiedKFold, LeaveOneOut, and a few other classes and functions from sklearn.model_selection.
A learning curve, sometimes called a training curve, shows how the prediction score of training and validation sets depends on the number of training samples. You can use learning_curve() to get this dependency, which can help you find the optimal size of the training set, choose hyperparameters, compare models, and so on.
Hyperparameter tuning, also called hyperparameter optimization, is the process of determining the best set of hyperparameters to define your machine learning model. sklearn.model_selection provides you with several options for this purpose, including GridSearchCV, RandomizedSearchCV, validation_curve(), and others. Splitting your data is also important for hyperparameter tuning.
Conclusion
You now know why and how to use train_test_split() from sklearn. You’ve learned that, for an unbiased estimation of the predictive performance of machine learning models, you should use data that hasn’t been used for model fitting. That’s why you need to split your dataset into training, test, and in some cases, validation subsets.
In this tutorial, you’ve learned how to:
- Use
train_test_split()to get training and test sets - Control the size of the subsets with the parameters
train_sizeandtest_size - Determine the randomness of your splits with the
random_stateparameter - Obtain stratified splits with the
stratifyparameter - Use
train_test_split()as a part of supervised machine learning procedures
You’ve also seen that the sklearn.model_selection module offers several other tools for model validation, including cross-validation, learning curves, and hyperparameter tuning.
If you have questions or comments, then please put them in the comment section below.
Get Your Code: Click here to download the free sample code that you’ll use to learn about splitting your dataset with scikit-learn’s train_test_split().
Take the Quiz: Test your knowledge with our interactive “Split Your Dataset With scikit-learn's train_test_split()” quiz. You’ll receive a score upon completion to help you track your learning progress:
Interactive Quiz
Split Your Dataset With scikit-learn's train_test_split()In this quiz, you'll test your understanding of how to use the train_test_split() function from the scikit-learn library to split your dataset into subsets for unbiased evaluation in machine learning.
Frequently Asked Questions
Now that you have some experience with scikit-learn’s train_test_split(), you can use the questions and answers below to check your understanding and recap what you’ve learned.
These FAQs are related to the most important concepts you’ve covered in this tutorial. Click the Show/Hide toggle beside each question to reveal the answer.
train_test_split() is a function from scikit-learn that you use to split your dataset into training and test subsets, which helps you perform unbiased model evaluation and validation.
x_train and y_train are the parts of your dataset that you use to train—or fit—your machine learning model. x_train contains the input data, while y_train contains the corresponding output labels.
When you set test_size=0.2 in train_test_split(), you specify that 20% of your dataset should be used as the test set for evaluating your model, with the remaining 80% used for training.
Yes, train_test_split() can handle imbalanced datasets by using the stratify parameter, which ensures that the class distribution in the training and test sets matches the original dataset.